
Tangible Arrays
Every building block in the Collection is at least 95% pure, as validated by NMR and LC-MS.
In modern drug discovery, biologically relevant compounds are usually generated from high-throughput screening or virtual screening For a new target, HTS might be the only way to identify bioactive compounds. However, for targets that are well known, retrieval of active compounds by screening tens of thousands to millions of structurally diverse compounds is neither economical nor efficient. Also searching for specific molecules through millions of structures in virtual databases is time consuming. To accelerate this process, we offer a Tangible Array Program.
Our customers select scaffold (fragment) based on their filter criteria and then we create a relatively small library containing all possible structural combinations. The client then can review these libraries and select specific compounds of his interest. This significantly speeds up the selection process.
Currently our collection covers diverse chemical and pharmacophoric space. Typically, to have good coverage and retain optimal chemical diversity, the size of a set for each selected scaffold varies from 100 to 700 compounds. We prepare these sets on scale, up to 100mg and at the designated purity.
Compounds can be offered on exclusive or non-exclusive bases.
Scaffolds Libraries can be:
- based on scaffolds of PBMR.
- based on scaffolds of the customer
- based on described procedures
Examples PBMR Scaffolds libraries:
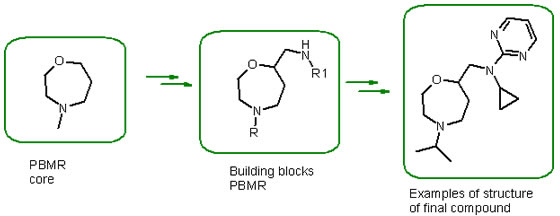
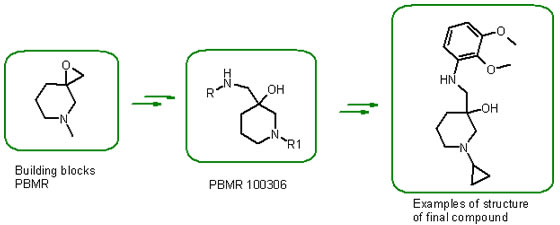
[1]Factors Determining the Selection of Organic Reactions by Medicinal Chemists and the Use of These Reactions in Arrays (Small Focused Libraries) Tony W. J. Cooper, Ian B. Campbell, Dr. Simon J. F. Macdonald /DOI: 10.1002/anie.201002238/Angewandte Chemie International Edition/ Volume 49, Issue 44, pages 8082–8091, October 25, 2010
[2] Generation of a Set of Simple, Interpretable ADMET Rules of ThumbM. Paul Gleeson*Computational and Structural Chemistry, GlaxoSmithKline Medicines Research Centre, Gunnels Wood Road, Stevenage, Hertfordshire, SG1 2NY, United Kingdom/J. Med. Chem., 2008, 51 (4), pp 817–834/DOI: 10.1021/jm701122q Publication Date (Web): January 31, 2008
[3] Lessons Learned from Molecular Scaffold Ye Hu, Dagmar Stumpfe, and Ju?rgen Bajorath*
Department of Life Science Informatics, B-IT, LIMES Program Unit Chemical Biology and Medicinal Chemistry, Rheinische Friedrich-Wilhelms-Universita?t, Dahlmannstr. 2, D-53113 Bonn, Germany J. Chem. Inf. Model., 2011, 51 (8), pp 1742–1753/ DOI: 10.1021/ci200179y
[4] The Medicinal Chemist’s Toolbox: An Analysis of Reactions Used in the Pursuit of Drug CandidatesStephen D. Roughley*‡ and Allan M. Jordan‡ Vernalis (R&D) Ltd., Granta Park, Cambridge, CB21 6GB, U.K.§ Cancer Research UK Drug Discovery Unit, Paterson Institute for Cancer Research, University of Manchester, Wilmslow Road, Manchester, M20 4BX, U.K. J. Med. Chem., 2011, 54 (10), pp 3451–3479/DOI: 10.1021/jm200187y
More information and examples can be found from the downloaded files:
New Products
-
Unique Organic Intermediates
Unique and popular organic BBL available in 24 hours shipping or synthesis on demand.
-
New Screening collection
Screening compounds from shelves in the client’s preferred format.
-
Tangible Arrays
Сarefully selected and designed based on biological and physicochemical properties of molecules.

